Search Results
Filter Options
| circRNA ID | Gene | Gene Type | Strand (circRNA/Gene) |
circRNA Type
The back-fusion junctions (BFJs) of circRNAs from genic regions may have three possible positions relative to the nearest exon-intron boundaries of their cognate linear transcripts, giving rise to three circRNA subtypes: (1) canonical (or completely boundary) circRNAs with both BFJs aligning to exon-intron boundaries; (2) half interior (or half boundary) circRNAs with one, but not both, of the BFJs aligning to exon-intron boundaries; (3) complete interior (or non-boundary) circRNAs with both junctions appearing inside of exons or introns. Lariats are byproducts of splicing and can be regarded as variants of canonical circRNAs; for clarity, we list lariats separately here. Intergenic circRNAs, derived from intergenic regions that often encode poorly annotated noncoding transcripts, can be considered a new distinct subtype of their own. We collectively refer to complete interior, half interior, and intergenic circRNAs as noncanonical circRNAs (nc-circRNAs) to distinguish them from canonical circRNAs (c-circRNAs). 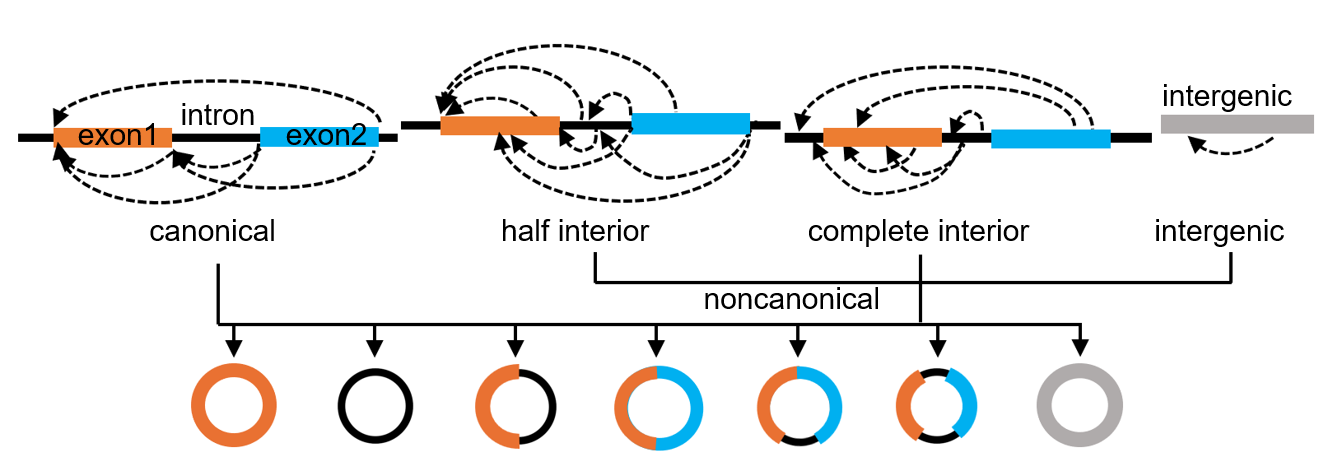
|
Position | Length | Species |
|---|---|---|---|---|---|---|---|
| MYL12B/18:63730737-63730745-63750032-63750040--/+ | MYL12B | protein_coding | -/+ | antisense | 18:63730744-63750039 | 19295.0 | Crab-eating macaque |
| GSTM5/1:113902534-113902535-113918959-113918960--/- | GSTM5 | protein_coding | -/- | partial | 1:113902535-113918960 | 16425.0 | Crab-eating macaque |
| PIGL/16:16159833-16159835-16160061-16160063--/- | PIGL | protein_coding | -/- | interior | 16:16159835-16160063 | 228.0 | Crab-eating macaque |
| intergenic/11:124264983-124264993-124264527-124264537-+/. | intergenic | Na | +/. | intergenic | 11:124264983-124264527 | 456.0 | Crab-eating macaque |
| intergenic/11:124264185-124264199-124264641-124264655--/. | intergenic | Na | -/. | intergenic | 11:124264186-124264642 | 456.0 | Crab-eating macaque |
| intergenic/11:124264983-124264993-124263841-124263851-+/. | intergenic | Na | +/. | intergenic | 11:124264983-124263841 | 1142.0 | Crab-eating macaque |
| intergenic/11:124264983-124264993-124263613-124263623-+/. | intergenic | Na | +/. | intergenic | 11:124264993-124263623 | 1370.0 | Crab-eating macaque |
| intergenic/11:124264392-124264412-124264620-124264640--/. | intergenic | Na | -/. | intergenic | 11:124264405-124264633 | 228.0 | Crab-eating macaque |
| intergenic/11:124264983-124264993-124264299-124264309-+/. | intergenic | Na | +/. | intergenic | 11:124264993-124264309 | 684.0 | Crab-eating macaque |
| CAPZB/1:205130593-205130597-205149570-205149574--/+ | CAPZB | protein_coding | -/+ | antisense | 1:205130597-205149574 | 18977.0 | Crab-eating macaque |