Search Results
Filter Options
| circRNA ID | Gene | Gene Type | Strand (circRNA/Gene) |
circRNA Type
The back-fusion junctions (BFJs) of circRNAs from genic regions may have three possible positions relative to the nearest exon-intron boundaries of their cognate linear transcripts, giving rise to three circRNA subtypes: (1) canonical (or completely boundary) circRNAs with both BFJs aligning to exon-intron boundaries; (2) half interior (or half boundary) circRNAs with one, but not both, of the BFJs aligning to exon-intron boundaries; (3) complete interior (or non-boundary) circRNAs with both junctions appearing inside of exons or introns. Lariats are byproducts of splicing and can be regarded as variants of canonical circRNAs; for clarity, we list lariats separately here. Intergenic circRNAs, derived from intergenic regions that often encode poorly annotated noncoding transcripts, can be considered a new distinct subtype of their own. We collectively refer to complete interior, half interior, and intergenic circRNAs as noncanonical circRNAs (nc-circRNAs) to distinguish them from canonical circRNAs (c-circRNAs). 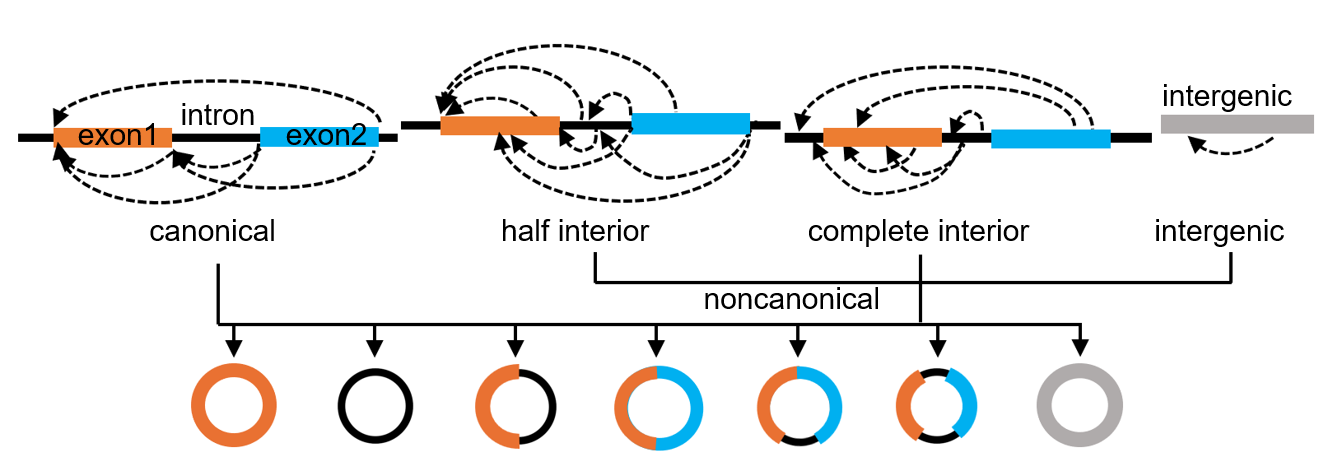
|
Position | Length | Species |
|---|---|---|---|---|---|---|---|
| intergenic/11:124264752-124264765-124264980-124264993--/. | intergenic | Na | -/. | intergenic | 11:124264752-124264980 | 228.0 | Crab-eating macaque |
| FAHD2A/13:446980-446982-469166-469168--/+ | FAHD2A | protein_coding | -/+ | antisense | 13:446982-469168 | 22186.0 | Crab-eating macaque |
| intergenic/11:124265013-124265027-124264101-124264115-+/. | intergenic | Na | +/. | intergenic | 11:124265013-124264101 | 912.0 | Crab-eating macaque |
| CAPZB/1:205147569-205147573-205163227-205163231--/+ | CAPZB | protein_coding | -/+ | antisense | 1:205147573-205163231 | 15658.0 | Crab-eating macaque |
| CCDC127::intergenic/6:305538-305540-351720-351722--/- | CCDC127::intergenic | protein_coding | -/- | intergenic | 6:305540-351722 | 46182.0 | Crab-eating macaque |
| intergenic::CCDC127/6:331616-331618-285705-285707-+/- | intergenic::CCDC127 | protein_coding | +/- | intergenic | 6:331616-285705 | 45911.0 | Crab-eating macaque |
| GSTM5/1:113949061-113949064-113937887-113937890-+/- | GSTM5 | protein_coding | +/- | antisense | 1:113949064-113937890 | 11174.0 | Crab-eating macaque |
| PIGL/16:16159521-16159526-16159977-16159982--/- | PIGL | protein_coding | -/- | interior | 16:16159521-16159977 | 456.0 | Crab-eating macaque |
| intergenic/11:124264222-124264229-124263994-124264001-+/. | intergenic | Na | +/. | intergenic | 11:124264229-124264001 | 228.0 | Crab-eating macaque |
| intergenic/11:124263928-124263933-124264842-124264847--/. | intergenic | Na | -/. | intergenic | 11:124263928-124264842 | 914.0 | Crab-eating macaque |